47 mIF距离分析
多重免疫荧光/空间蛋白组学切片染色的细胞空间距离分析
1. 文献中的一个“多重免疫荧光/空间蛋白组学切片染色的细胞空间距离分析”
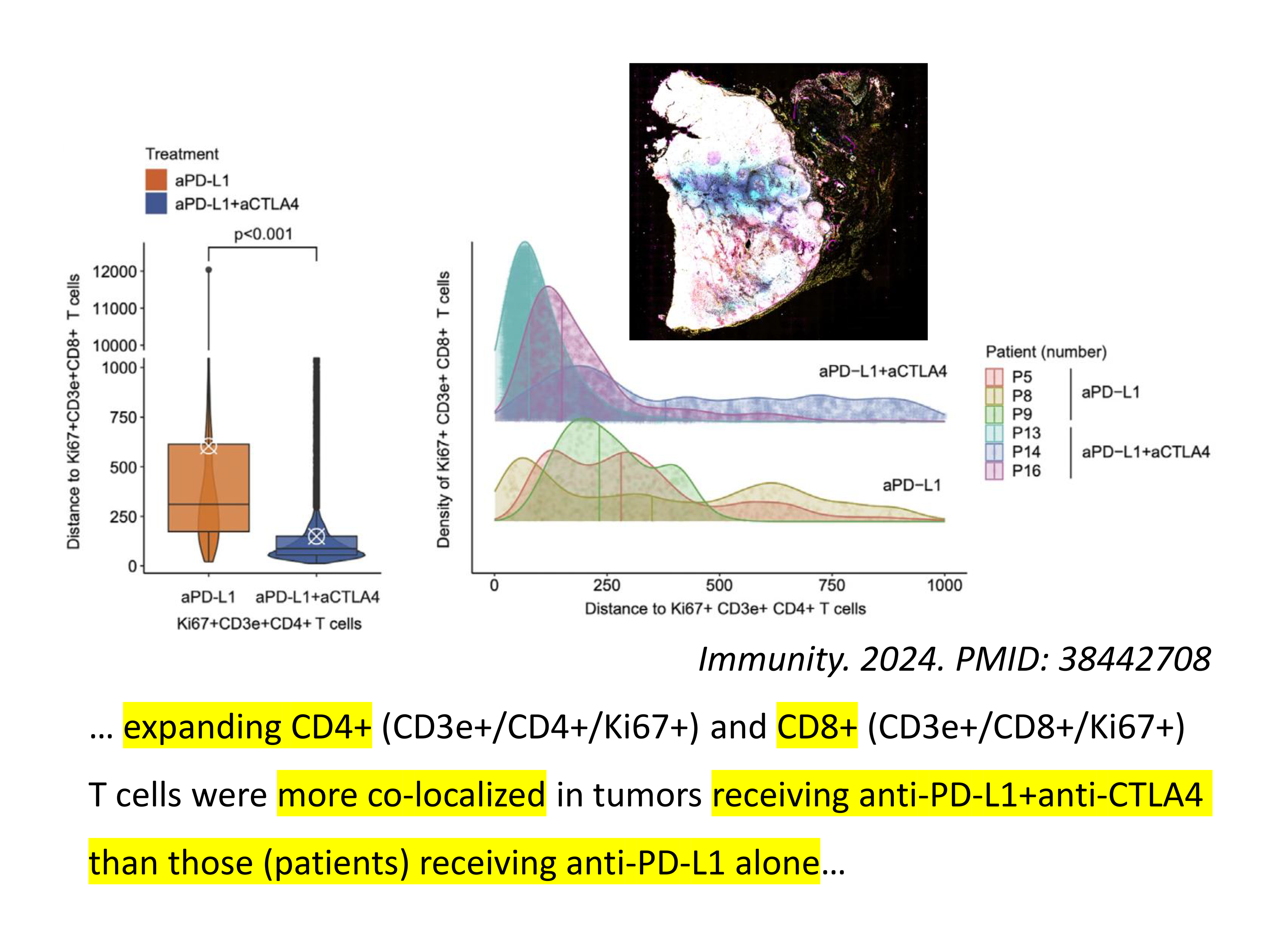
… expanding CD4+ (CD3e+/CD4+/Ki67+) and CD8+ (CD3e+/CD8+/Ki67+) T cells were more co-localized in tumors receiving anti-PD-L1+anti-CTLA4 than those (patients) receiving anti-PD-L1 alone … (1)👋
… 扩展的CD4+(CD3e+/CD4+/Ki67+)和CD8+(CD3e+/CD8+/Ki67+)T细胞在接受anti-PD-L1+anti-CTLA4治疗的肿瘤中比单独接受anti-PD-L1治疗的肿瘤中有更多的共定位 …(1)👋
该文献作者就患者肿瘤组织的多重免疫荧光染色/空间蛋白质组学确实发现了联合治疗组(anti-PD-L1+anti-CTLA1)中,CD4+ T细胞和CD8+ T细胞的空间距离更近(相比于anti-PD-L1治疗)(1)。确实是一个有意义的发现🤩。
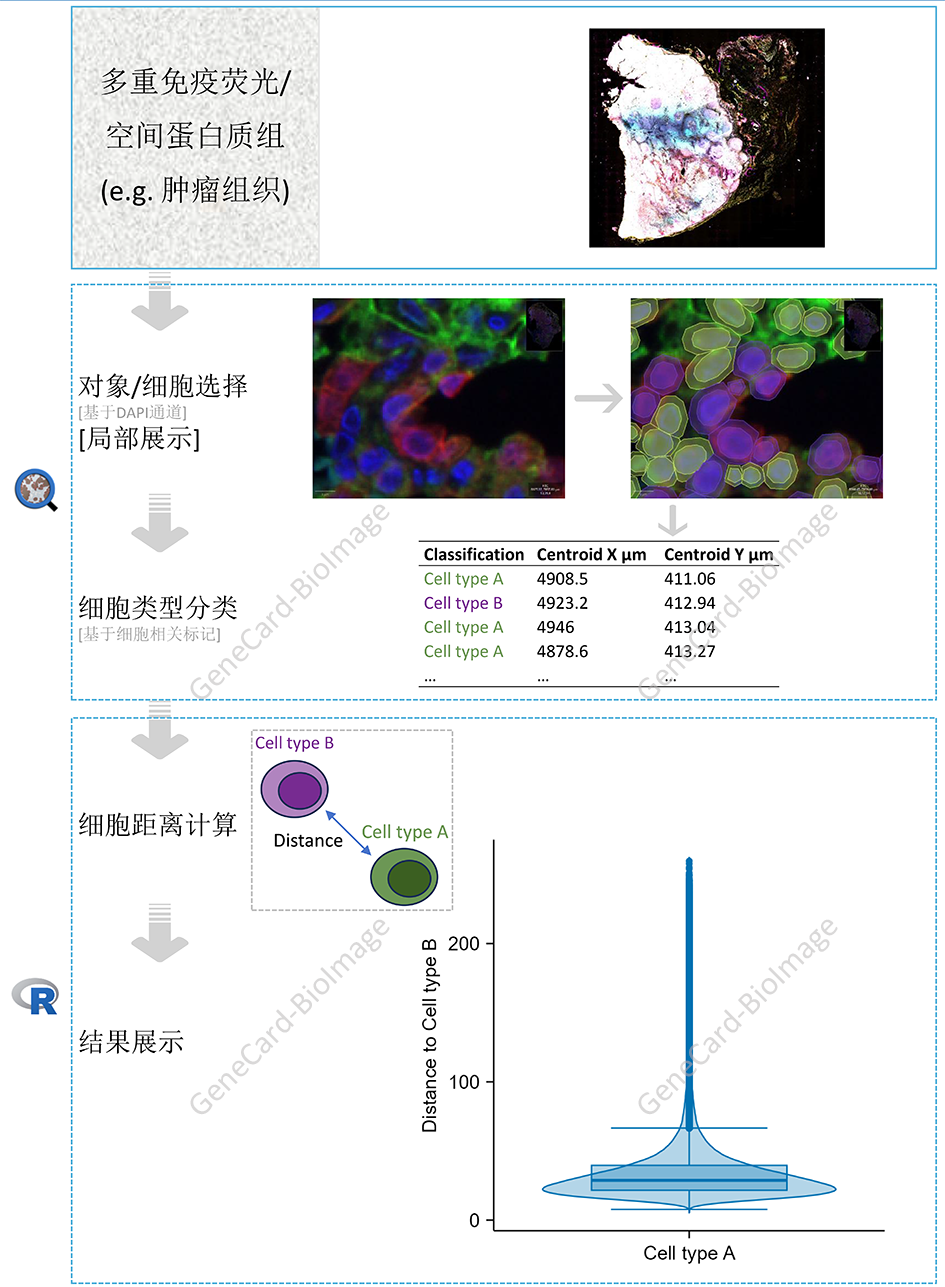
实话实说/to be honest😎,该文献作者亦公布了其分析的步骤(2),该步骤整体上用到了QuPath软件(包括1个深度学习扩展工具)(3) (4)和R语言(包括另外3个R包)(5)。
今卡-生物图像花了些时间和精力(无法评估算多还是算少😃)了解文献作者的分析思路,并复现了该分析步骤,及做了些许修改。
2. 分析步骤示意图:多重免疫荧光/空间蛋白组学切片染色的细胞空间距离分析
References
1.
A. Franken, M. Bila, A. Mechels, S. Kint, J. Van Dessel, V. Pomella, S. Vanuytven, G. Philips, O. Bricard, J. Xiong, B. Boeckx, S. Hatse, T. Van Brussel, R. Schepers, C. Van Aerde, S. Geurs, V. Vandecaveye, E. Hauben, V. Vander Poorten, S. Verbandt, K. Vandereyken, J. Qian, S. Tejpar, T. Voet, P. M. Clement, D. Lambrechts, CD4+ t cell activation distinguishes response to anti-PD-L1+anti-CTLA4 therapy from anti-PD-L1 monotherapy. Immunity 57, 541–558.e7 (2024).
2.
A. Franken, M. Bila, D. Lambrechts, Protocol for whole-slide image analysis of human multiplexed tumor tissues using QuPath and r. STAR Protocols 5, 103270 (2024).
3.
P. Bankhead, M. B. Loughrey, J. A. Fernández, Y. Dombrowski, D. G. McArt, P. D. Dunne, S. McQuaid, R. T. Gray, L. J. Murray, H. G. Coleman, J. A. James, M. Salto-Tellez, P. W. Hamilton, QuPath: Open source software for digital pathology image analysis. Scientific Reports 7 (2017).
4.
M. P. Humphries, P. Maxwell, M. Salto-Tellez, QuPath: The global impact of an open source digital pathology system. Computational and Structural Biotechnology Journal 19, 852–859 (2021).
5.
R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2025; https://www.R-project.org/).