20 QuPath的Omnipose
QuPath的Omnipose插件的安装和运行
相对来说,能成功的安装Omnipose (1)(或者包括Cellpose (2))软件,并成功的安装和运行对应QuPah插件,相对困难些。
这里展示在Windows 11 64位操作系统的安装过程。
1. 安装Omnipose
假设已经安装好了miniconda(本机安装的版本是conda 25.9.1)。
参照https://github.com/kevinjohncutler/omnipose安装Omnipose。
conda create -n omnipose 'python==3.10.12' pytorch
conda activate omnipose
git clone https://github.com/kevinjohncutler/omnipose.git
cd omnipose
pip install -e .安装结束后,启动Omnipose遇到问题😥:
omnipose
OMP: Error #15: Initializing libiomp5md.dll, but found libiomp5md.dll already initialized.
OMP: Hint This means that multiple copies of the OpenMP runtime have been linked into the program. That is dangerous, since it can degrade performance or cause incorrect results. The best thing to do is to ensure that only a single OpenMP runtime is linked into the process, e.g. by avoiding static linking of the OpenMP runtime in any library. As an unsafe, unsupported, undocumented workaround you can set the environment variable KMP_DUPLICATE_LIB_OK=TRUE to allow the program to continue to execute, but that may cause crashes or silently produce incorrect results. For more information, please see http://www.intel.com/software/products/support/.遇到了OpenMP and libiomp5相关问题,参考https://omnipose.readthedocs.io/installation.html,运行:
conda install nomkl再次启动Omnipose,又遇到问题😥:
omnipose
Traceback (most recent call last):
File "C:\ProgramData\miniconda3\envs\omnipose\lib\runpy.py", line 196, in _run_module_as_main
return _run_code(code, main_globals, None,
File "C:\ProgramData\miniconda3\envs\omnipose\lib\runpy.py", line 86, in _run_code
exec(code, run_globals)
File "C:\ProgramData\miniconda3\envs\omnipose\Scripts\omnipose.exe\__main__.py", line 2, in <module>
File "D:\8_tak\36_Omnipose-study_2025-10-31\omnipose\src\omnipose\__init__.py", line 15, in <module>
from numba.core.errors import NumbaPendingDeprecationWarning
File "C:\ProgramData\miniconda3\envs\omnipose\lib\site-packages\numba\__init__.py", line 59, in <module>
_ensure_critical_deps()
File "C:\ProgramData\miniconda3\envs\omnipose\lib\site-packages\numba\__init__.py", line 34, in _ensure_critical_deps
import numpy as np
ModuleNotFoundError: No module named 'numpy'因此:
pip install numpy安装的numpy版本为v2.2.6。
这时,可以成功的启动Omnipose了🥳。
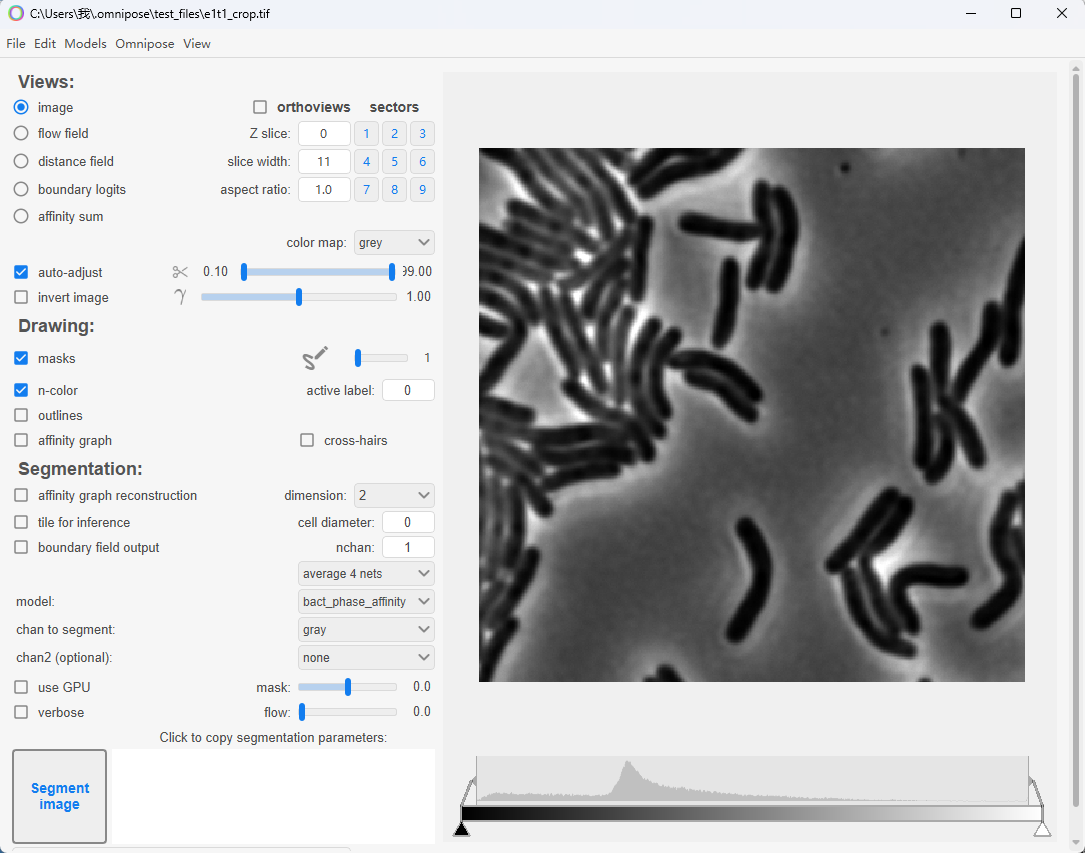
2. 安装QuPath的Omnipose插件
参考https://github.com/BIOP/qupath-extension-cellpose安装Omnipose插件并设置对应环境Python的路径,这里不再赘述。
运行插件提供的脚本(认真阅读脚本的注释,并根据需要去修改)时,报错(下面是部分报错信息)😥:
INFO: Cellpose2D: AttributeError: `newbyteorder` was removed from the ndarray class in NumPy 2.0. Use `arr.view(arr.dtype.newbyteorder(order))` instead.
INFO: Reading Temp_0_0_z0_t0_cp_masks.tif
ERROR: Cannot invoke "ij.ImagePlus.getProcessor()" because "label_imp" is null因此:
conda activate omnipose
pip install "numpy<2.0"安装的numpy版本降低为v1.26.4(第1部分安装的是v2.2.6)。
这时,可以成功的运行Omnipose插件了🥳。
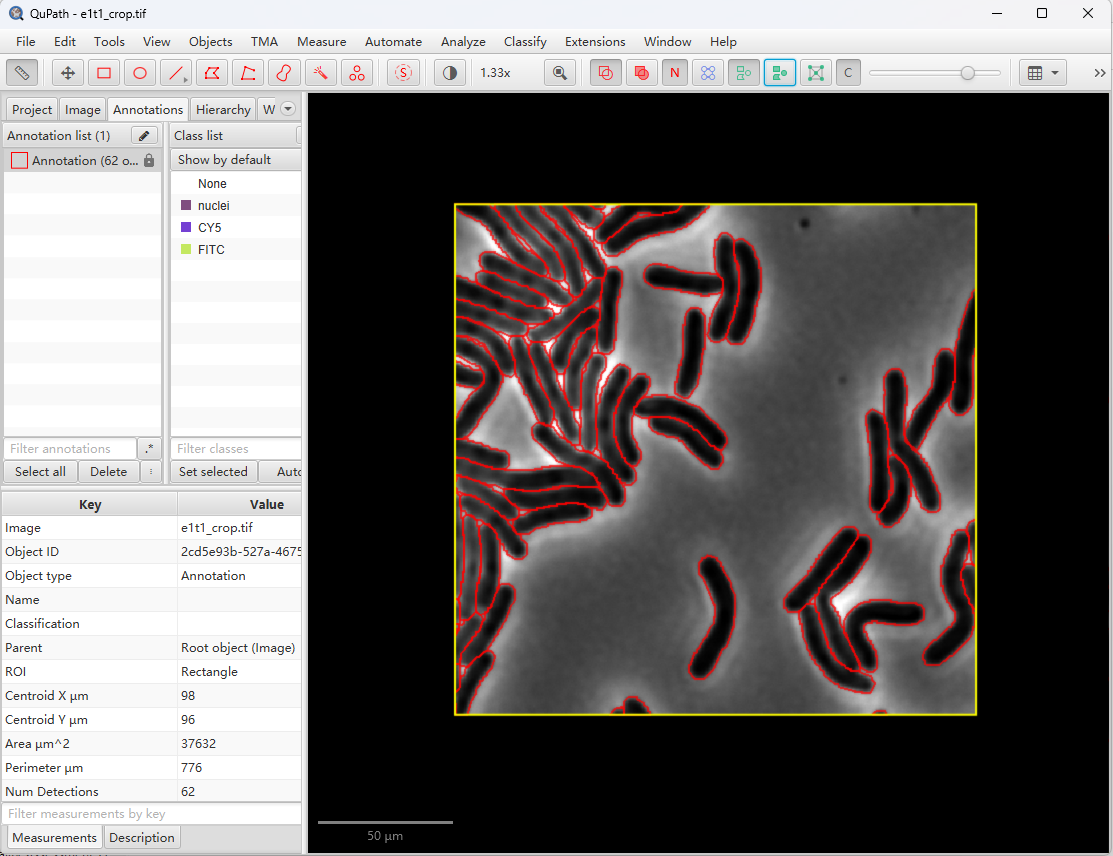
补充说明:脚本中的model用绝对路径去定位,比如:
def pathModel = "C:\\Users\\我\\.cellpose\\models\\bact_phase_affinitytorch_0"以防出现如下报错:
ERROR: Cannot invoke "ij.ImagePlus.getProcessor()" because "label_imp" is nullReferences
1.
K. J. Cutler, C. Stringer, T. W. Lo, L. Rappez, N. Stroustrup, S. Brook Peterson, P. A. Wiggins, J. D. Mougous, Omnipose: A high-precision morphology-independent solution for bacterial cell segmentation. Nature Methods 19, 1438–1448 (2022).
2.
M. Pachitariu, M. Rariden, C. Stringer, Cellpose-SAM: Superhuman generalization for cellular segmentation. doi: 10.1101/2025.04.28.651001 (2025).